Hello!
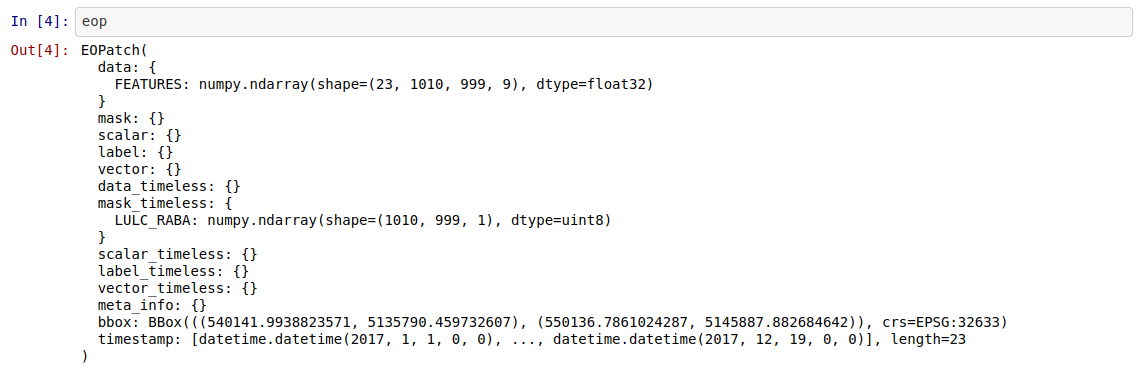
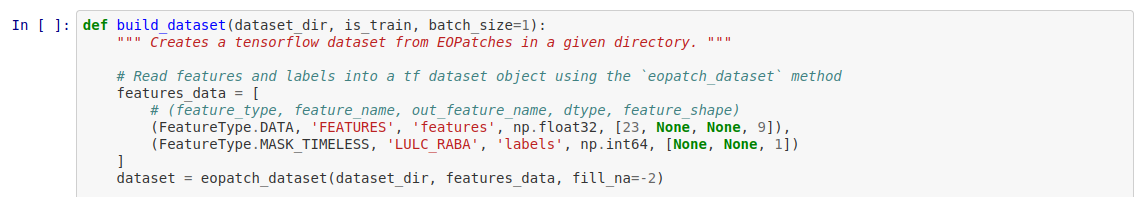
I am trying to run through the examples notebook from the EO-Flow repository and I am having an issue that will not allow me to run through the entire notebook. Down where we begin to prepare the input data we have some import statements from EO-Flow but I keep getting a module not found error. I have tried most of what I could think of and what I found online but I cannot get past this point. If anyone can provide me some assistance I would greatly appreciate it.