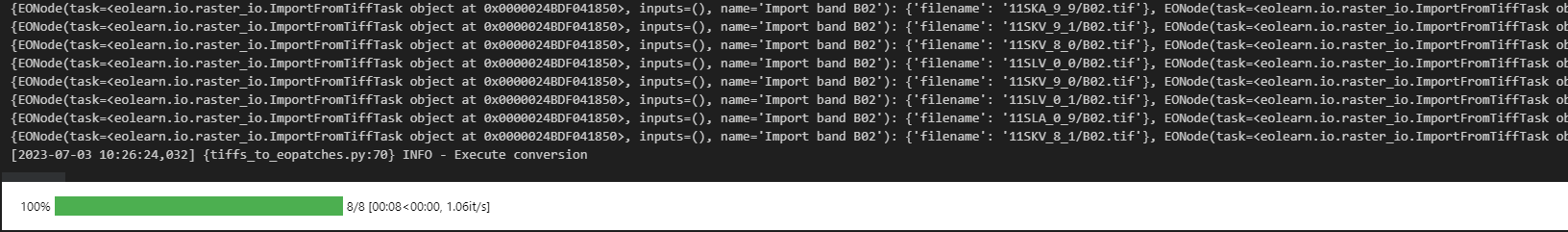
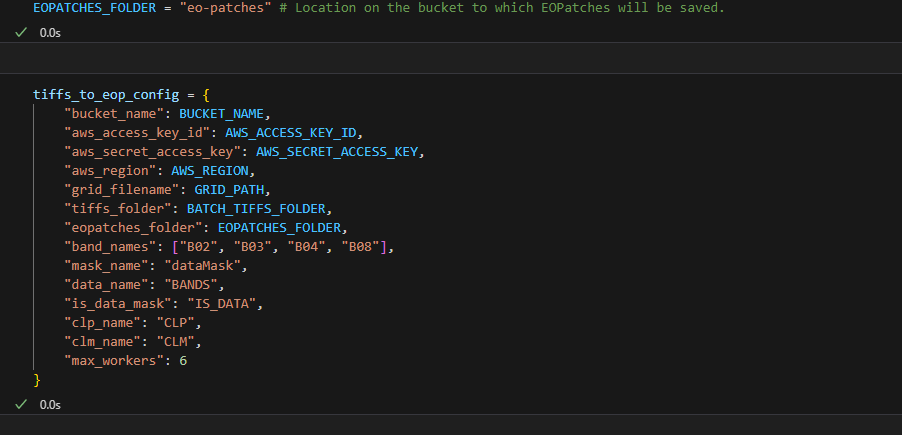
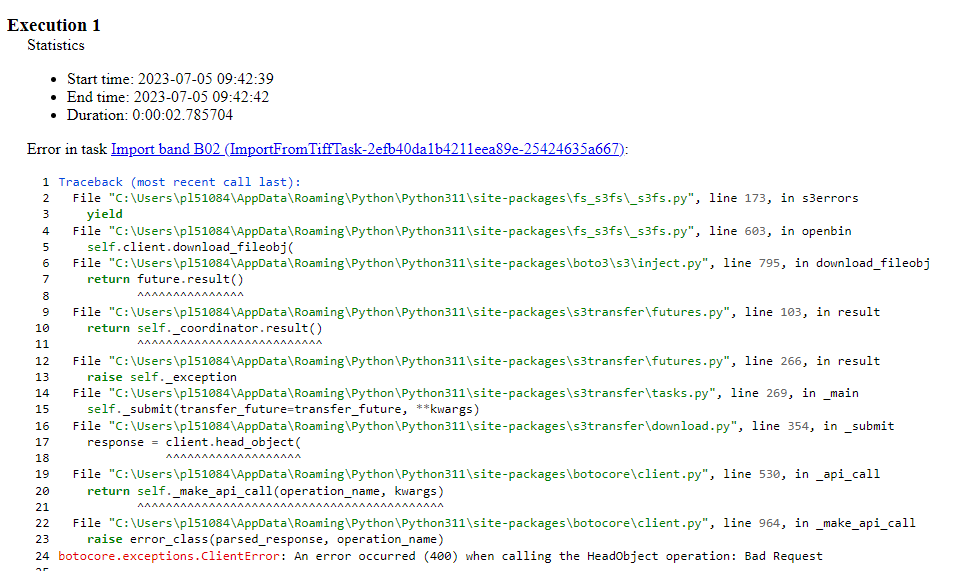
I am repurposing the following code from the field-delineation GitHub:
def get_tiffs_to_eopatches_workflow(config: TiffsToEopatchConfig, delete_tiffs: bool = False) -> EOWorkflow:
""" Set up workflow to ingest tiff files into EOPatches """
# Set up credentials in sh config
sh_config = set_sh_config(config)
import_bands = [(ImportFromTiffTask((FeatureType.DATA, band),
folder=f's3://{config.bucket_name}/{config.tiffs_folder}',
config=sh_config), f'Import band {band}')
for band in config.band_names]
import_clp = (ImportFromTiffTask((FeatureType.DATA, config.clp_name),
folder=f's3://{config.bucket_name}/{config.tiffs_folder}',
config=sh_config), f'Import {config.clp_name}')
import_mask = (ImportFromTiffTask((FeatureType.MASK, config.mask_name),
folder=f's3://{config.bucket_name}/{config.tiffs_folder}',
config=sh_config), f'Import {config.mask_name}')
rearrange_bands = (RearrangeBands(), 'Swap time and band axis')
add_timestamps = (AddTimestampsUpdateTime(f's3://{config.bucket_name}/{config.tiffs_folder}'), 'Load timestamps')
merge_bands = (MergeFeatureTask(
input_features={FeatureType.DATA: config.band_names},
output_feature=(FeatureType.DATA, config.data_name)), 'Merge band features')
remove_bands = (RemoveFeatureTask(features={FeatureType.DATA: config.band_names}), 'Remove bands')
rename_mask = (RenameFeatureTask((FeatureType.MASK, config.mask_name, config.is_data_mask)), 'Rename is data mask')
calculate_clm = (CloudMasking(), 'Get CLM mask from CLP')
save_task = (SaveTask(path=f's3://{config.bucket_name}/{config.eopatches_folder}', config=sh_config,
overwrite_permission=OverwritePermission.OVERWRITE_FEATURES), 'Save EOPatch')
filenames = [f'{band}.tif' for band in config.band_names] + \
[f'{config.mask_name}.tif', f'{config.clp_name}.tif', 'userdata.json']
delete_files = (DeleteFiles(path=config.tiffs_folder, filenames=filenames), 'Delete batch files')
workflow = [*import_bands,
import_clp,
import_mask,
rearrange_bands,
add_timestamps,
merge_bands,
remove_bands,
rename_mask,
calculate_clm,
save_task]
if delete_tiffs:
workflow.append(delete_files)
return linearly_connect_tasks(*workflow) #LinearWorkflow(*workflow)
Would linearly_connect_tasks function the same way as the old LinearWorkflow function? I can’t find any old documentation regarding LinearWorkflow