Hi SenHub-Team,
I tried the eoLearn-slovenia Land Cover Classification script.
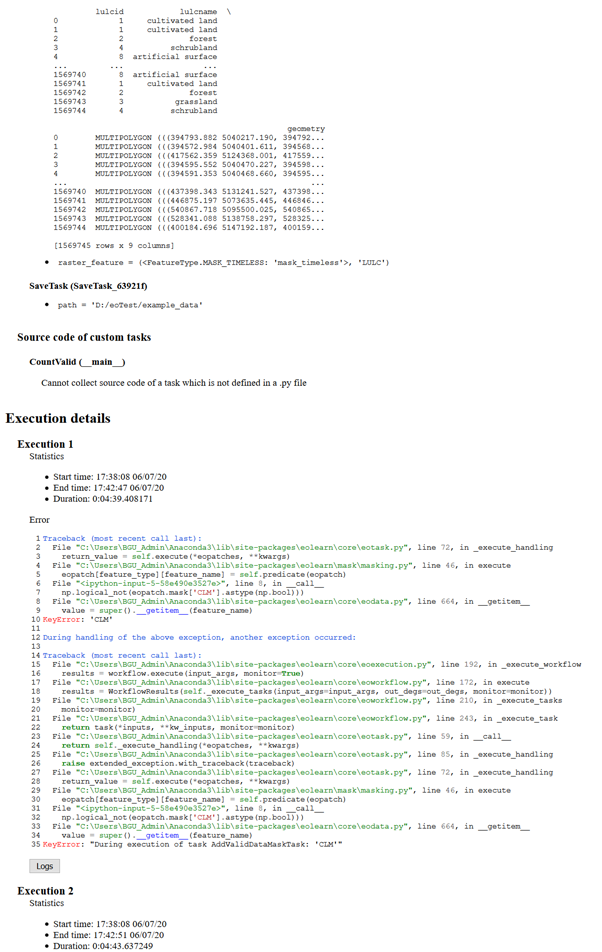
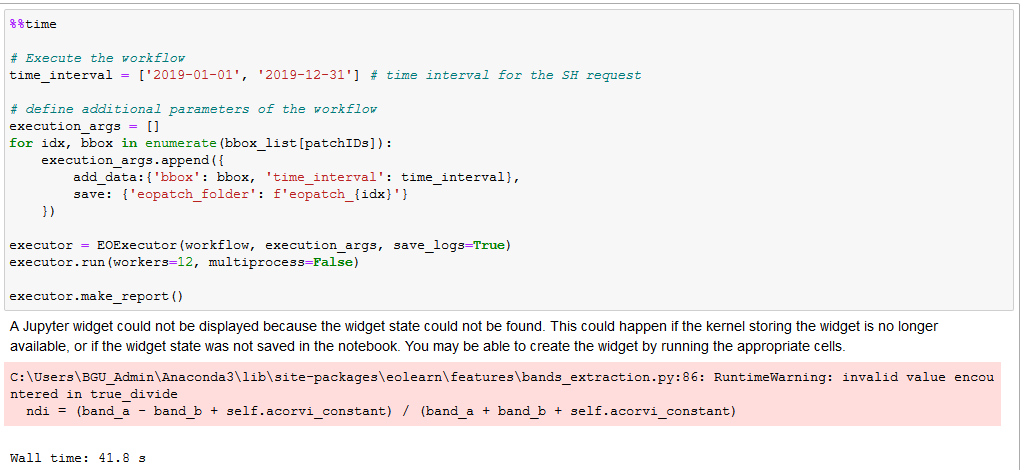
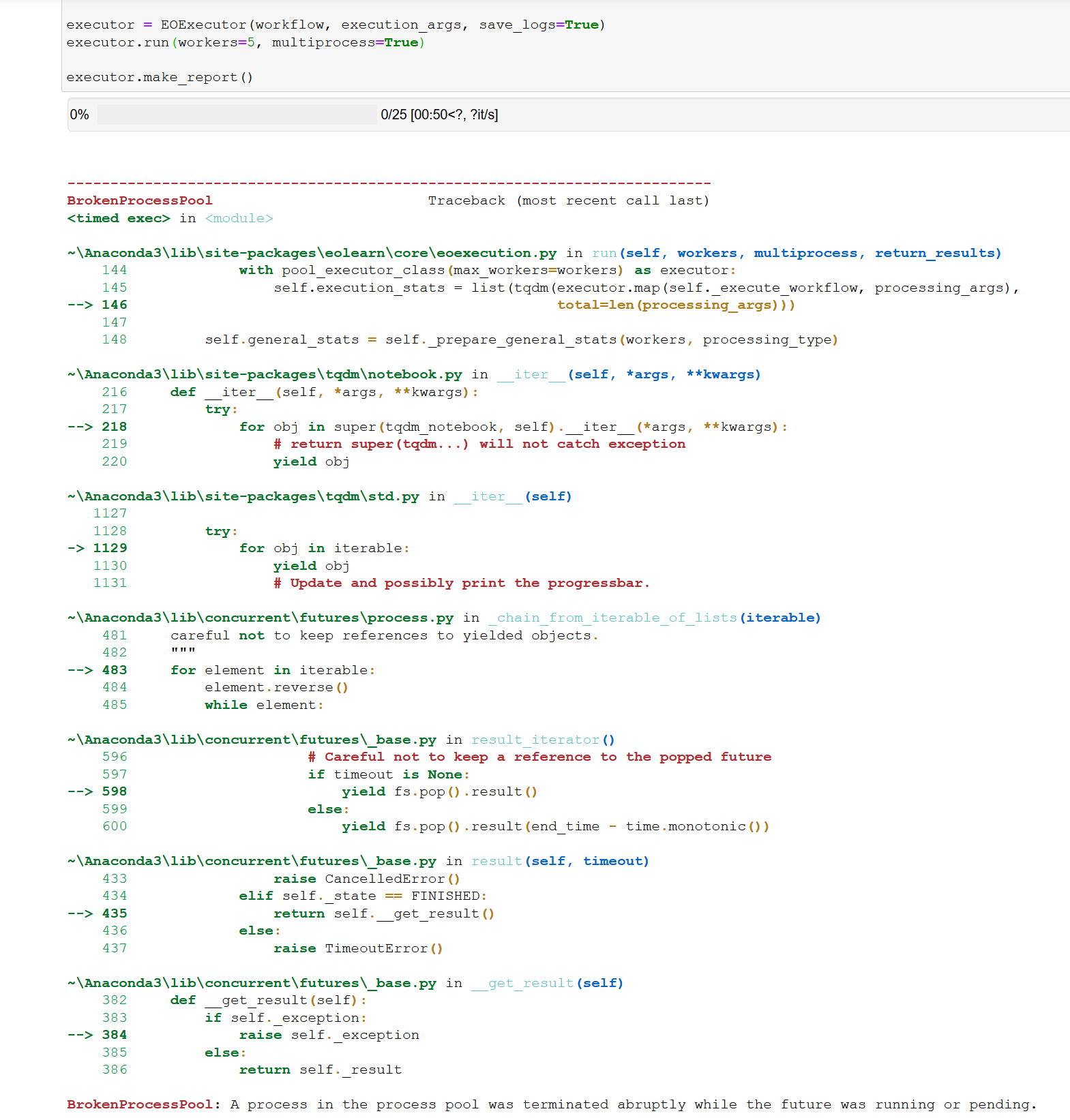
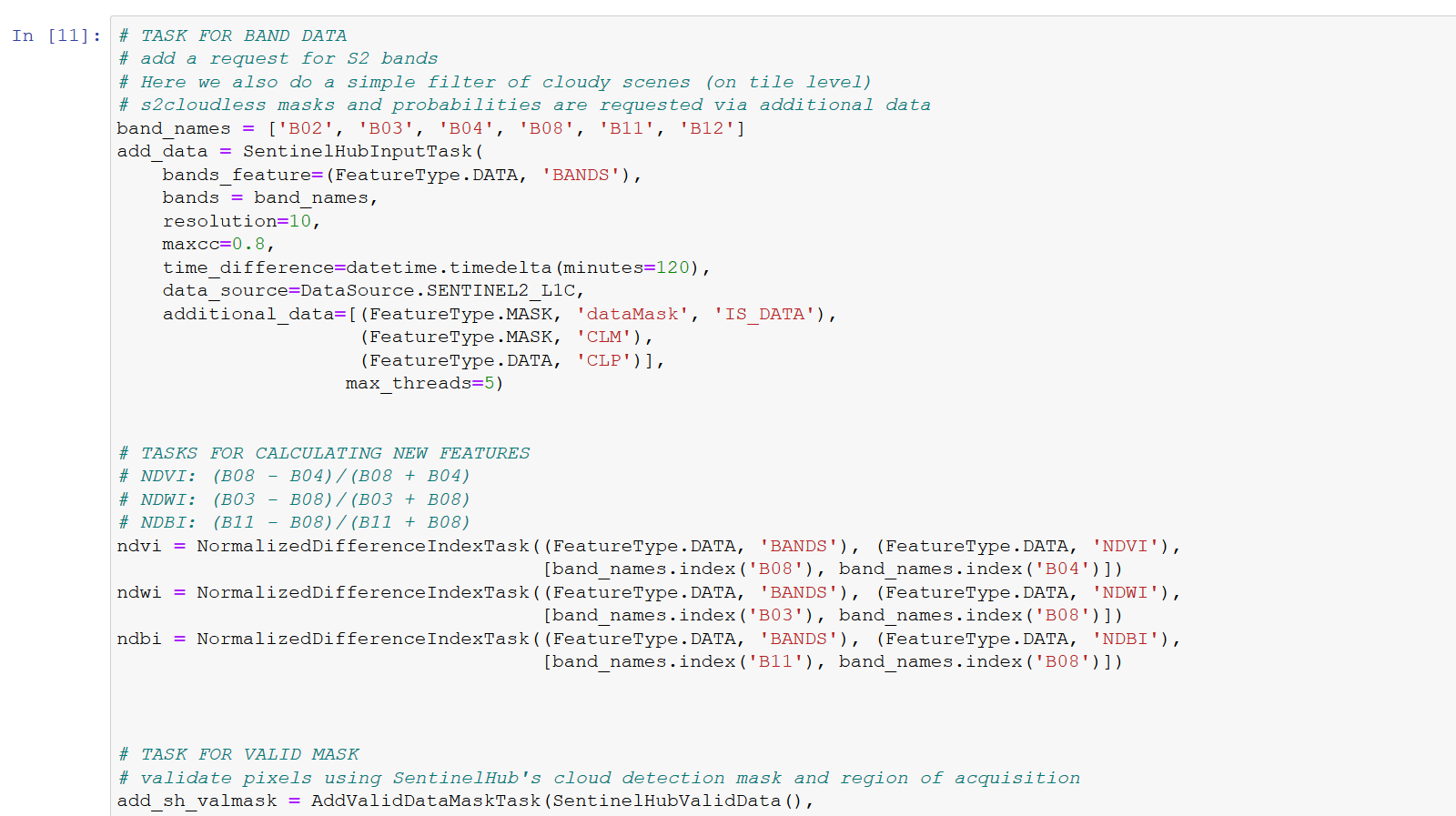
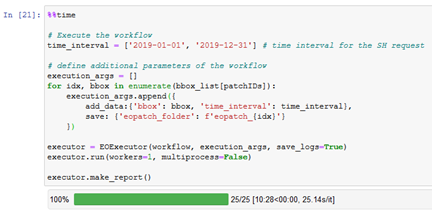
I have a problem with download the patches. Once it download the data for slovenia. After that I get only error messages for slovenia and in this case my area:
It has Problems with the CLM.
Has something change after?:
https://shforum.sinergise.com/t/cloud-masks-available-as-clm-clp-band/2113
What can I do?
Thank you very much for your work.
Greetz