Hi!
I am quite new to the EO space, so apologies if this is a dumb or simple question! I also realize I’m working off an example notebook that is no longer updated, so I understand if this issue cannot be solved. Anyway, I posted this in the eo-learn Github yesterday but figured I’d drop it here as well in case the hivemind could lend me a hand:
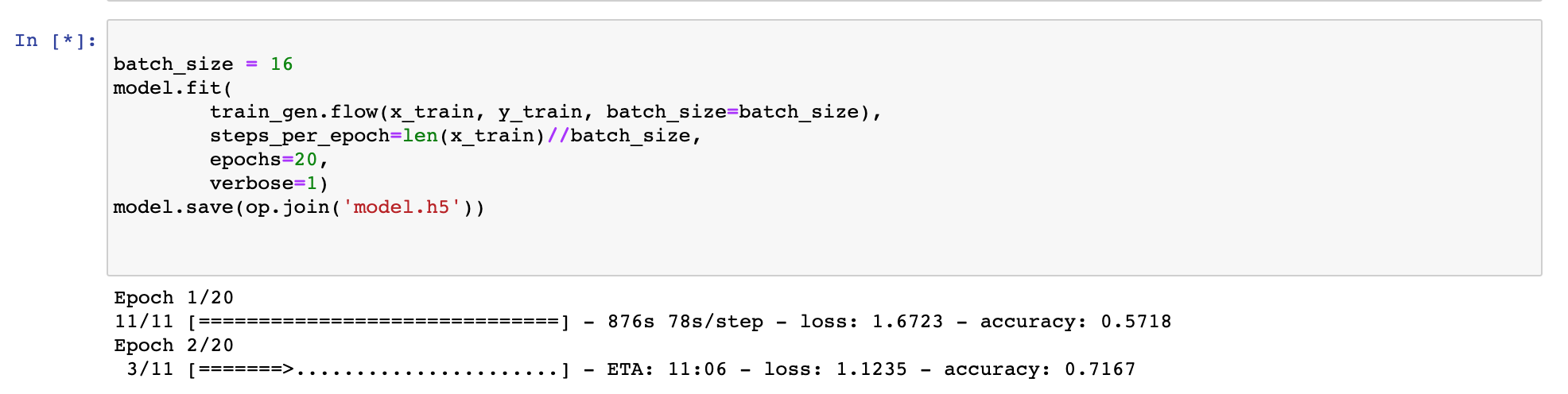
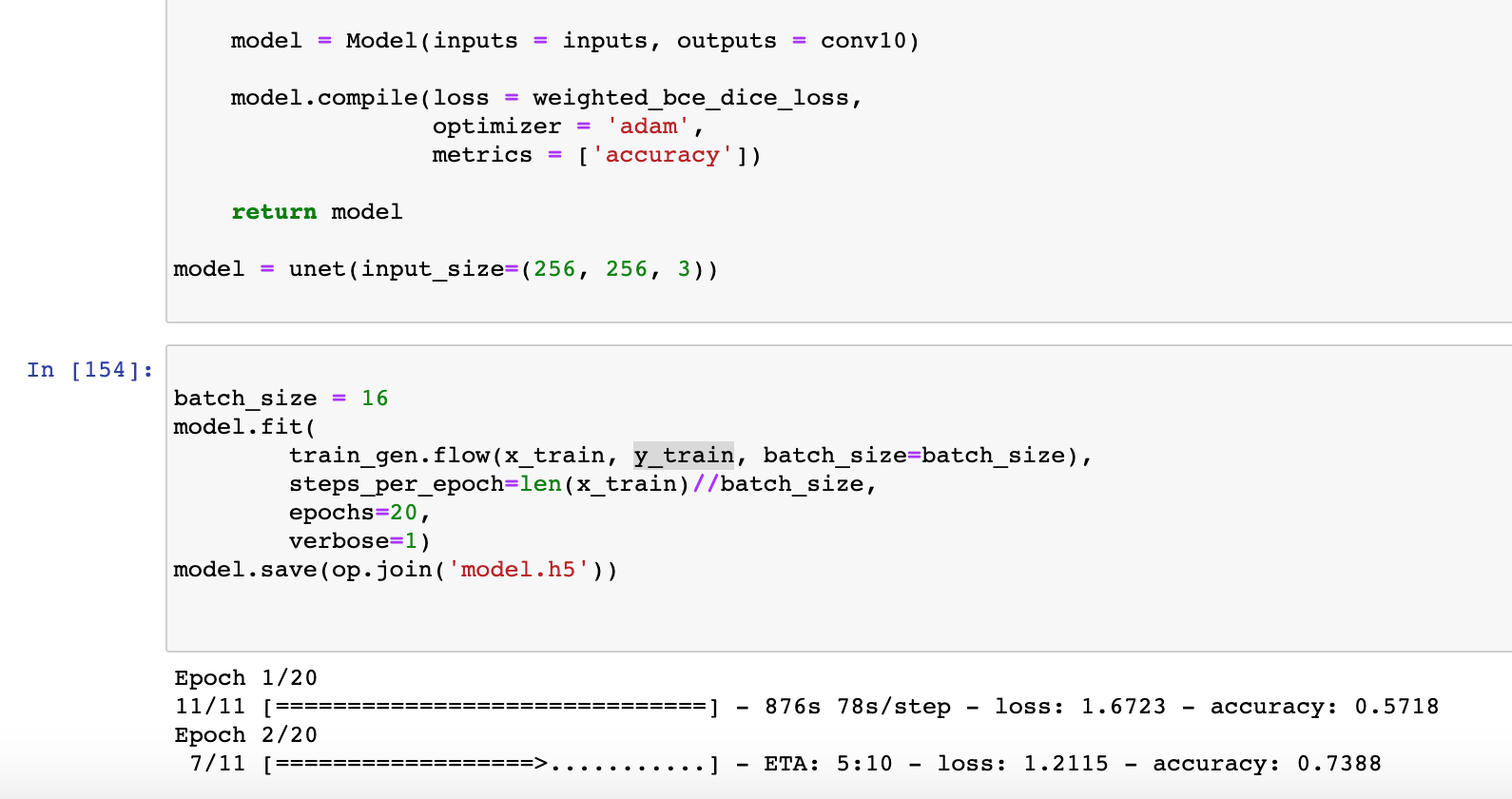
When I run up to Cell 9 in this notebook, I get the error:
“ValueError: During execution of task AddGeopediaFeature: operands could not be broadcast together with shapes (640,779,3) (4,)”
I’ve looked through the forums and existing issues/questions but haven’t found one that addresses the differing operands. I also tried the task without the MASK_TIMELESS feature and the Sentinel 2 imagery import worked fine (but obviously that is not the point of the example notebook).
The other thing that could be at the root is that the ground truth data I’m using is the EU Tree Cover Geopedia data, while my AOI (the paradise.geojson file) is over northern California. Some screenshots are attached - any help resolving this would be greatly appreciated and once again, apologies if it’s a silly question 🙂 Here’s the code in my Jupyter Notebook:
import os
import datetime
from os import path as op
import itertools
from eolearn.io import *
from eolearn.core import EOTask, EOPatch, LinearWorkflow, FeatureType, SaveTask, OverwritePermission
from sentinelhub import BBox, CRS, BBoxSplitter, MimeType, ServiceType, DataCollection
from tqdm import tqdm_notebook as tqdm
import matplotlib.pyplot as plt
import numpy as np
import geopandas
from sklearn.metrics import confusion_matrix
from keras.preprocessing.image import ImageDataGenerator
from keras import backend as K
from keras.models import *
from keras.layers import *
from keras.optimizers import *
from keras.utils.np_utils import to_categorical
K.clear_session()
from sentinelhub import SHConfig
config = SHConfig()
config.sh_client_id = “client id here”
config.sh_client_secret = “secret id here”
config.save()
time_interval = (‘2019-01-01’, ‘2019-12-31’)
img_width = 256
img_height = 256
maxcc = 0.2
get the AOI and split into bboxes
crs = CRS.UTM_10N
aoi = geopandas.read_file(‘path to paradise.geojson here’)
aoi = aoi.to_crs(crs=crs.pyproj_crs())
aoi_shape = aoi.geometry.values[-1]
bbox_splitter = BBoxSplitter([aoi_shape], crs, (19, 10))
aoi.plot()
aoi_width = aoi_shape.bounds[2] - aoi_shape.bounds[0]
aoi_height = aoi_shape.bounds[3] - aoi_shape.bounds[1]
print(f’Dimension of the area is {aoi_width:.0f} x {aoi_height:.0f} m2’)
set raster_value conversions for our Geopedia task
see more about how to do this here:
raster_value = {
‘0%’: (0, [0, 0, 0, 0]),
‘10%’: (1, [163, 235, 153, 255]),
‘30%’: (2, [119, 195, 118, 255]),
‘50%’: (3, [85, 160, 89, 255]),
‘70%’: (4, [58, 130, 64, 255]),
‘90%’: (5, [36, 103, 44, 255])
}
import matplotlib as mpl
tree_cmap = mpl.colors.ListedColormap([’#F0F0F0’,
‘#A2EB9B’,
‘#77C277’,
‘#539F5B’,
‘#388141’,
‘#226528’])
tree_cmap.set_over(‘white’)
tree_cmap.set_under(‘white’)
bounds = np.arange(-0.5, 6, 1).tolist()
tree_norm = mpl.colors.BoundaryNorm(bounds, tree_cmap.N)
create a task for calculating a median pixel value
class MedianPixel(EOTask):
“”"
The task returns a pixelwise median value from a time-series and stores the results in a
timeless data array.
“”"
def init(self, feature, feature_out):
self.feature_type, self.feature_name = next(self._parse_features(feature)())
self.feature_type_out, self.feature_name_out = next(self._parse_features(feature_out)())
def execute(self, eopatch):
eopatch.add_feature(self.feature_type_out, self.feature_name_out,
np.median(eopatch[self.feature_type][self.feature_name], axis=0))
return eopatch
initialize tasks
task to get S2 L2A images
input_task = SentinelHubInputTask(data_collection=DataCollection.SENTINEL2_L2A,
bands_feature=(FeatureType.DATA, ‘BANDS’),
resolution=10,
maxcc=0.2,
bands=[‘B04’, ‘B03’, ‘B02’],
time_difference=datetime.timedelta(hours=2),
additional_data=[(FeatureType.MASK, ‘dataMask’, ‘IS_DATA’)]
)
geopedia_data = AddGeopediaFeature((FeatureType.MASK_TIMELESS, ‘TREE_COVER’),
layer=‘ttl2275’, theme=‘QP’, raster_value=raster_value)
task to compute median values
get_median_pixel = MedianPixel((FeatureType.DATA, ‘BANDS’),
feature_out=(FeatureType.DATA_TIMELESS, ‘MEDIAN_PIXEL’))
task to save to disk
save = SaveTask(op.join(‘data’, ‘eopatch’),
overwrite_permission=OverwritePermission.OVERWRITE_PATCH,
compress_level=2)
initialize workflow
workflow = LinearWorkflow(input_task, geopedia_data, get_median_pixel, save)
use a function to run this workflow on a single bbox
def execute_workflow(index):
bbox = bbox_splitter.bbox_list[index]
info = bbox_splitter.info_list[index]
patch_name = 'eopatch_{0}_row-{1}_col-{2}'.format(index,
info['index_x'],
info['index_y'])
results = workflow.execute({input_task:{'bbox':bbox, 'time_interval':time_interval},
save:{'eopatch_folder':patch_name}
})
return list(results.values())[-1]
del results
idx = 10
example_patch = execute_workflow(idx)