Hi, I have downloaded RapidEye spectral reflectance image analytic products for 2011, 2013, 2015, 2017, and 2019. No color balancing was applied to create a seamless color because it may alter the original reflectance value. Now, if you look at the attached images from 2013, it looks like there is a binary color difference. Also, there is a square shape in the 2011 images. If I use these images for biomass modeling, will this color difference cause a problem or error? If so, how can it be fixed
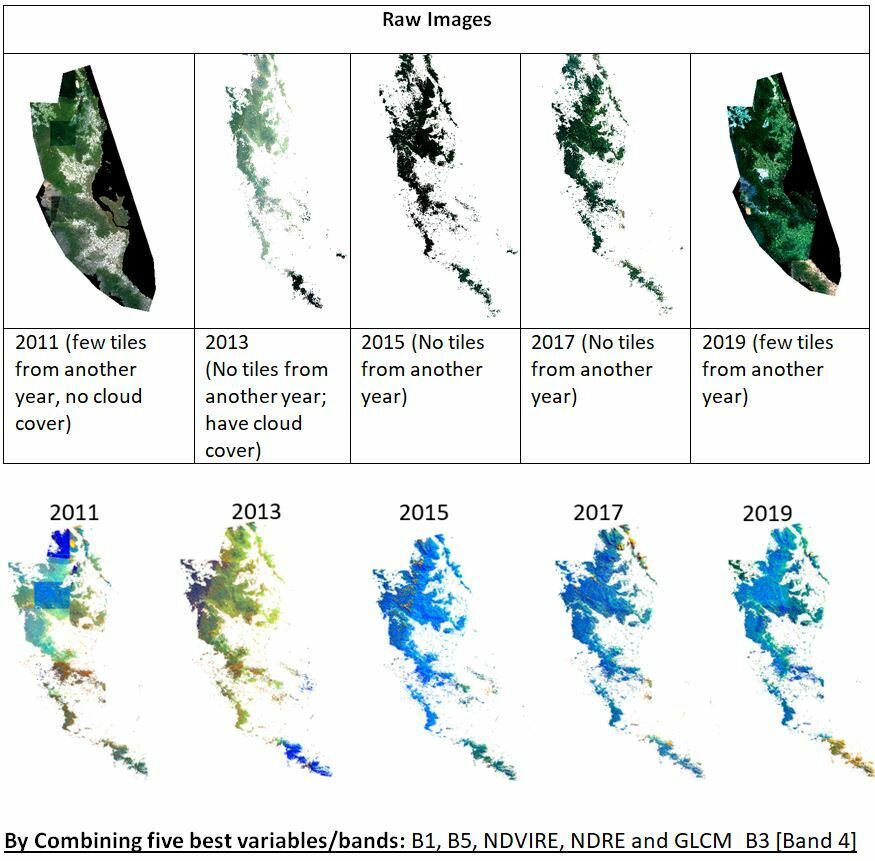
in QGIS?
Any suggestions in this matter are highly appreciated.
Thank you.





